Introduction
Overview
Teaching: 0 min
Exercises: 0 minQuestions
What is Bioconductor and why would I use it?
Objectives
Describe what kinds of resources Bioconductor provides and give an example of what they might be useful for.
Locate Bioconductor packages and learn their intended purpose.
Locate Bioconductor package reference manuals and vignettes.
Install Bioconductor packages using biocLite.
What is Bioconductor?
Bioconductor is a collection of open-source tools that use R. It contains over 1400 R packages that can be used to analyze many types of biological data. A major benefit of Bioconductor is that the packages are usually well-documented, and there is an active community of users and developers.
Bioconductor Packages
There are three types of Bioconductor packages:
- Software: packages of code that can be used to analyze data
- Annotations: packages that contain useful information like genome sequences or transcript coordinates
- Experiments: packages that contain experimental data, usually used to provide examples and test software
Bioconductor packages are available for many different steps in data analysis pipelines. Often, you can choose between several different Bioconductor packages for the same step in the pipeline. This allows you to try different approaches and pick which one is the best fit for your project or data.
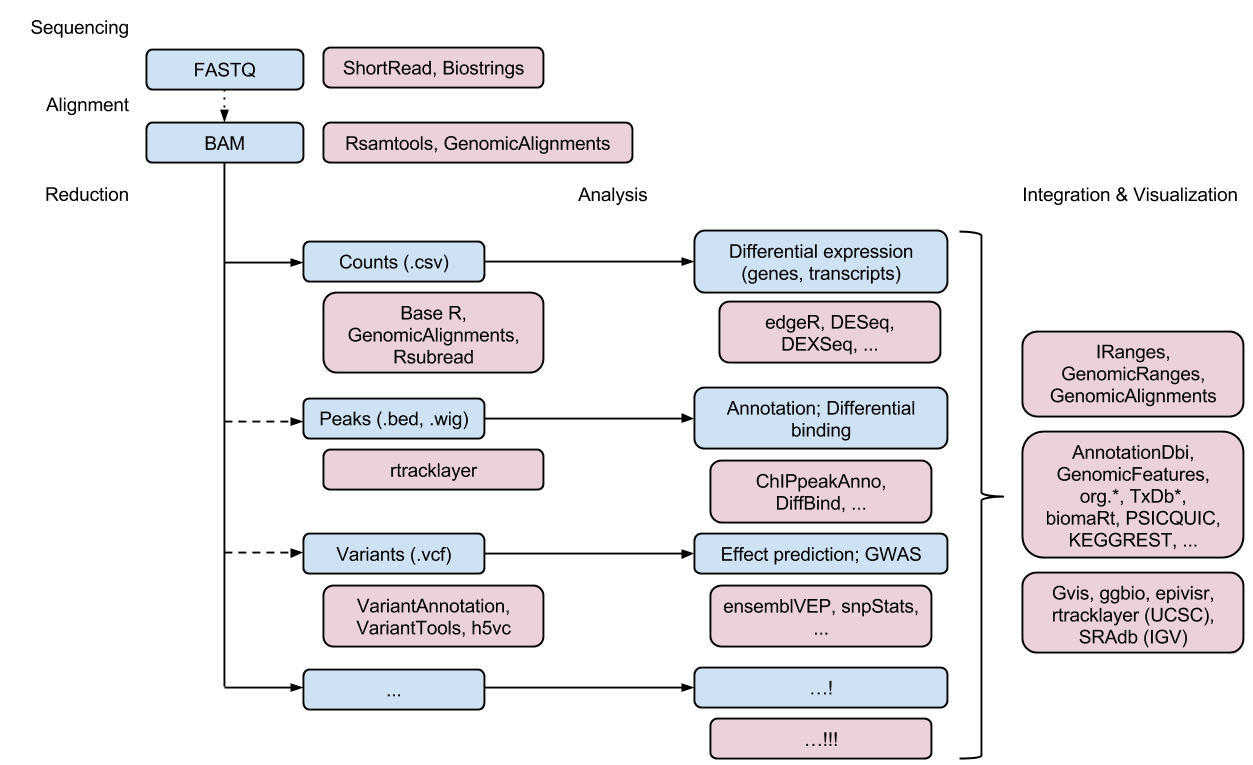 Image from https://www.bioconductor.org/help/course-materials/2017/OSU/B1_Bioconductor_intro.html
Image from https://www.bioconductor.org/help/course-materials/2017/OSU/B1_Bioconductor_intro.html
Exploring Bioconductor Packages
A browsable list of Bioconductor packages is available in biocViews on the Bioconductor website
Take a look at some of the packages available. Later in this lesson, we’ll be looking for differential expression of genes in RNA-Seq data. Find a Bioconductor package that could be useful for that.
Solution
By browsing to Software, then BiologicalQuestion, then DifferentialExpression you’ll see 241 packages available, from ABSSeq to xps.
Bioconductor Documentation
One of the best things about Bioconductor is the documentation available for its packages. Packages have (at least) both a Reference Manual and a Vignette. The Reference Manual describes the package in a standardized way, indicating its usage and listing the arguments available. The Vignette is more like a tutorial: it provides the information you need to get started, and takes you through some example workflows.
Finding Reference Manuals and Vignettes
For example, take a look at the biocViews page for the popular differential gene expression analysis package DESeq2.
Under “Documentation” you’ll see links to both the Vignette (called Analyzing RNA-Seq data with DESeq2) and the Reference Manual.
Install Bioconductor
As with any R package, in order to use a Bioconductor package you must first install it. There is a special method of installing Bioconductor packages, called biocLite, that is different from installing other R packages. To get started, first you need to get biocLite. To do so, run this command in RStudio:
source("https://bioconductor.org/biocLite.R")
biocLite()
Now you can use BiocLite to install Bioconductor packages. BiocLite helps to manage updates and make sure that Bioconductor packages can work together (almost) seamlessly.
Key Points
Bioconductor is one source for biological data analysis tools.
Bioconductor packages are documented with reference manuals and vignettes
Bioconductor packages are installed with biocLite